2. Basic Model Setup and Run
This exercise/tutorial walks through the process of making a basic model simulation. With this exercise you will get to interact with the code and a bunch of the supporting tools, including Docker, and Git. For post processing and visualization, see the Plotting Example.
Hint
Quick-start summary
For users who have Docker and Git installed and some familiarity with the command line.
Note
These instructions should work for users with Podman rather than Docker, but several additional steps may be necessary to ensure that Podman can run with non-root access. See this issue for more information.
The settings shown here will use the input data that is shipped with the code
in the repository’s demo-data/ directory. The run will output a single
variable (GPP), and will run for 2 pixels.
Only the steps are described here. For quick-start instructions with example commands see the README.
Clone the repository and change into the directory:
Make yourself locations for input data and your modeling workflows. We don’t actually use the input directory for this quick-start, but creating it and setting the
envvariable below prevents some warnings. On some systems you will need to make Docker usable by non-root users. See here.Build Docker images. For this demo we only build 2 of the images. For your needs you might want to build the other images. See
./docker-build-wrapper.sh --helpfor more info.Setup your environment variables
V_TAG,DDT_INPUT_CATALOG, andDDT_WORKFLOWS. You can do this in a.envfile, your.bashrc, or by exporting them to your shell. For example using the.envfile.Start the Docker containers.
Obtain a shell in the container.
Compile the code.
Setup a working directory for your model run and change into it. The
setup_working_directory.pyscript bootstraps a new directory to hold your model run. This includes a few configuration files and a folder for the outputs.Adjust run mask, config, outputs as necessary. The default settings should run 1 or 2 pixels and produce outputs for GPP. There are more tools provided in the
scripts/directory for working with the config file, run mask, output specifications and parameters.Start the model run.
Visualize run.
Note
This lab is more or less a duplicate of an existing wiki page: https://github.com/uaf-arctic-eco-modeling/dvm-dos-tem/wiki/How-To:-Run-dvmdostem-and-plot-output-via-Docker.
We are in the process of migrating all content from the wiki and assorted Google Documents to this Sphinx based documentation system, therefore this document should be the most up-to-date.
This tutorial will walk you through the process of:
getting the
dvmdostemcode,building your Docker images,
starting your Docker containers, and
running
dvmdostemin your Docker container (in a trivial fashion).
This tutorial will walk you through most of the above steps with varying levels of hand-holding.
If you need more background help, please see the Prelude document which has information on Version Control, Docker, and general programming.
2.1. Install tools
Get started by installing all the software you will need.
2.1.1. Version control
We are using version control software called Git to manage our code base. Git is a distributed version control system. The software works well for managing contributions to the code base from a lot of people, although this is not “free” - it still takes quite a bit of learning, practice and work to make this happen smoothly. Git allows you to track changes to files, to make branches for isolating groups of changes and to manage merging changes from different lines of development.
In addition to Git, we are using the code hosting service Github. Git is a stand alone version control system, whereas Github is a hosting service that integrates with Git. Github is a cloud based code hosting service that facilitates collaboration between developers without the need to host and manage a server. In addition to basic code hosting Github provides a heap of additional features including issue tracking, access control, and various continuous integration, testing and publishing tools.
For more general information and help about Git, see the Prelude - Version Control section.
For more information and details about how the dvmdostem project is using
Git, please see the Dev Info - Version
Management section.
For more information about how the dvmdostem project is using Github, see
the Using Github
Features
document.
Git terms you should familiarize yourself with before continuing: commit, branch, merge, tag, repository, remote.
Github terms you should familiarize yourself with before continuing: clone, fork, release, wiki, issue tracker, pull request.
2.1.2. Run time environment and virtualization with Docker
Any software, dvmdostem included, requires a specific set of hardware and
supporting software in order to run. This is referred to as the “runtime
environment”. There may be additional tools needed for compiling the software
(translating the software from source code to machine instructions) and for
working with the code to improve it (developing the code). We are currently
using a containerization tool called Docker to simplify and standardize the
creation of these environments. Docker helps to create stand alone, self
sufficient containers in which an application can run. The idea is that a Docker
container should be portable and able to run on a wide variety of host systems.
The dependencies for a piece of software are isolated inside the container. This
isolation allows software with conflicting dependencies to run on the host
system.
Hint
These instructions should work for users with Podman rather than Docker, but several additional steps may be necessary to ensure that Podman can run with non-root access. See this issue for more information.
There are other ways you can get the environment necessary for running,
compiling and developing dvmdostem, such as a native installation, or using
a virtual machine (e.g. with Vagrant and Virtual Box or VMWare). Each path to a
functioning runtime environment has its tradeoffs and can be useful in different
situations. We have successfully used native installs, a VirtualBox VM with
Vagrant, and Docker to achieve a valid runtime for dvmdostem. This tutorial
is based on Docker, but the directions for building a VM using Vagrant are still
included in the virtualmachines/ directory of the repository. You may notice
that both the Docker approach and the Vagrant/VirtualBox VM approach basically
build off of a generic Ubuntu base system and then simply install a variety of
packages, adjust some settings and manage sharing data between the host and
guest system.
Our recent switch to using Docker does not preclude you from pursuing a native install or a virtual machine, but Docker provides several advantages, namely that the containers are smaller and lighter weight than a full virtual machine and that the internal layout of the container is the same for everyone, so paths and other settings can be shared between developers.
Docker also represents a paradigm shift that can take some getting used to - in
fact we are still working on how dvmdostem should best fit within the
paradigm. With Docker the concept is to isolate a single process and its
dependencies into a container. The container is then run as a service; ideally
there is one process per container and the process offers a single service. Not
all work naturally fits into this paradigm and we expect to modify the
dvmdostem Docker stack in the near future as we improve how things are
structured. See this Note for more
information.
Note
One way to think of Docker is to imagine that you have an office with several old computers laying around. And you have a system you want to build that requires a few different computers, each with slightly different software installed and running. And for your system, these different computers will need to talk to each other and share certain data. To assemble and configure each computer, you have a CD with the basic operating system you need, i.e. a Windows install CD, one for Linux, and another for a different flavor of Linux with some special packages installed. Once you install the operating system on each of your computers, you can start the computer and leave it running so it can talk to the other computers once you get them setup. In a notebook, you write down the steps for each installation and other settings to get the computer running and connected with the shared drives for communication. With this analogy, the Docker images are analogous to the CDs you have. Docker containers are analogous to the running instances of the computers. And docker-compose is analogous to the instructions you wrote in the paper notebook for starting the whole system.
Docker terms you should familiarize yourself with before continuing: build, image, container, volume, docker-compose.
2.1.3. Text editors and terminal emulator
You will also need a text editor that you will use to view and modify files and some kind of terminal emulator (shell or console program) on your computer. As of 2022 popular text editors are Sublime, VSCode, and Atom. MacOS and Linux generally have an easily accessible terminal program. For Windows, look into MobaXTerm.
2.1.4. Summary
So to get going, do the following if you do not already have these tools:
Install a text editor and terminal program.
Install Git on your computer. Directions for this vary based on your operating system; you should be able to get started here https://git-scm.com. When you are done you should be able to run
git --version.Install Docker. Again directions for this vary for your operating system but you should be able to get started here https://docs.docker.com. When you are done, you should be able to open a terminal and run
$ docker infoand$ docker --versionand get something like this:$ docker info Client: Context: default Debug Mode: false Plugins: buildx: Docker Buildx (Docker Inc., v0.7.1) compose: Docker Compose (Docker Inc., v2.2.1) scan: Docker Scan (Docker Inc., v0.14.0) Server: Containers: 4 ...much more info below... $ docker --version Docker version 20.10.11, build dea9396
2.2. Get the code
With your tools setup, it is time to get the source code. Navigate to https://github.com/uaf-arctic-eco-modeling/dvm-dos-tem and find the link to clone the repository.
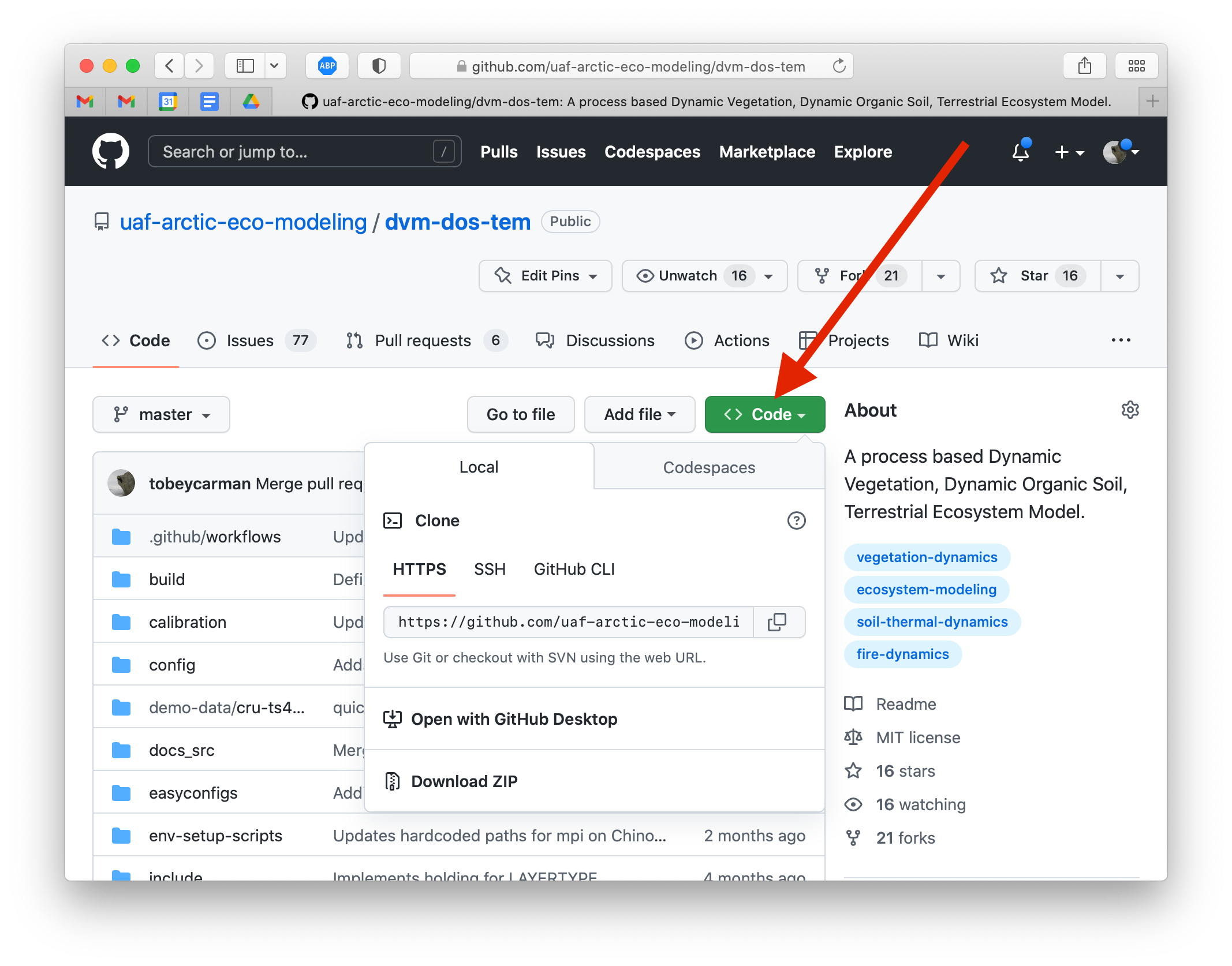
On your computer, open a terminal and navigate to a place where you would like
your copy of dvmdostem to be stored. Copy the clone address and use it to
run the $ git clone command in your terminal.
Note
Using ssh vs https clone address. Notice that the “clone” button on
github gives you the option to use either the https address (default) or
the ssh address. If you are have been added to the project as a
collaborator you should use the ssh address so that you are able to push
changes to the upstream fork. If you use the https address, you will still
be able to push to your personal fork, but will not be able to push to the
upstream uaf-arctic-eco-modelling fork.
You might notice that the clone address is simply the URL for the repo with
.git at the end. This will fetch a copy of the repository from Github to
your local machine. You should see some messages in your terminal to that
extent. Notice that on your machine you now have a new directory entitled
dvm-dos-tem with an exact copy of the code that is on Github. In addition,
due to the power of Git, you also have the entire history of the project on your
computer as well. This works because inside your dvm-dos-tem/ directory is
another (hidden) folder named .git - this hidden folder contains the history
of the project and all the other information that Git needs to perform its
magic. You rarely, if ever, need to look at the contents of the .git
directory. Take a few minutes to explore the files in the dvm-dos-tem
directory.
Note
Sometimes we write dvmdostem, sometimes we write dvm-dos-tem and
sometimes we write DVM-DOS-TEM. These are all the same thing. The order is
always the same, but sometimes we use capitals and sometimes lower case,
sometimes with hyphens and sometimes without. This is a fluke of history. In
some cases it looks better capitalized, sometimes it looks better lower case.
The repository ended up with hyphens in the name, but the compiled binary
executable does not have hyphens.
Warning
Notice that when you run $ git remote -v you are presented with some text
indicating that your remote is named ‘origin’ and points to the Github
uaf-arctic-eco-modeling repository. To be consistent with this tutorial and
the rest of our documentation, you should rename this remote to ‘upstream’ and
point ‘origin’ to your personal fork of the code (if you have one). To do this
use the $ git remote rename <old> <new> command.
Warning
Notice that after cloning the repository and running the $ git branch
command you are on the master branch of the code. It is highly recommended
that you set up your terminal so that the git branch is displayed in your
prompt. The directions for this are terminal/shell specific and widely
available with a little web searching. A decent example for Ubuntu/bash can be
found here:
https://askubuntu.com/questions/730754/how-do-i-show-the-git-branch-with-colours-in-bash-prompt.
2.3. Build Docker images
Now that you have the code on your machine, you need a way to interact with it. You can browse the files using standard tools on your computer, but to execute (run) the code you will need a special environment with all the dependencies installed. This is where Docker comes into play.
With Docker there are two steps to using the software: building the images and
starting the running containers based on the images. As of dvmdostem v0.6.0,
there are 5 images that we are using for this project:
cpp-dev- general C++ development tooling.
dvmdostem-dev- all tools necessary for developing and working withdvmdostem; this will be the image that most users will use most of the time. Relies on mounted volumes for access to the source code.
dvmdostem-build- a stripped down image only used for compiling the C++ portion of the code. Includes the source code inside the image instead of relying on mounted volumes.
dvmdostem-run- a very small production image with only the necessary run-time libraries, no development or compiling tooling.
dvmdostem-mapping-support- an image with GDAL tools installed and Python.
Note
In the (hopefully near) future it should not be necessary to build your own images unless you have very specific development needs. The images will be automatically built and published (to Github, maybe elsewhere) by Github Actions with each release of the code.
With the existing layout, images 1-4 are successively built on top of each other
(layered) which allows for faster builds when you only need to re-build because
of a change in something in one of the upper layers. The
dvmdostem-mapping-support image is totally separate from the others and
allows installing GDAL which is difficult to do in conjunction with some of the
libraries that dvmdostem depends on.
To build your images, you can use the docker-build-wrapper.sh script. You
should examine the commands and comments in this script as well as the
Dockerfile in order to understand what is going on. If the wrapper script fails,
you can try running each step individually.
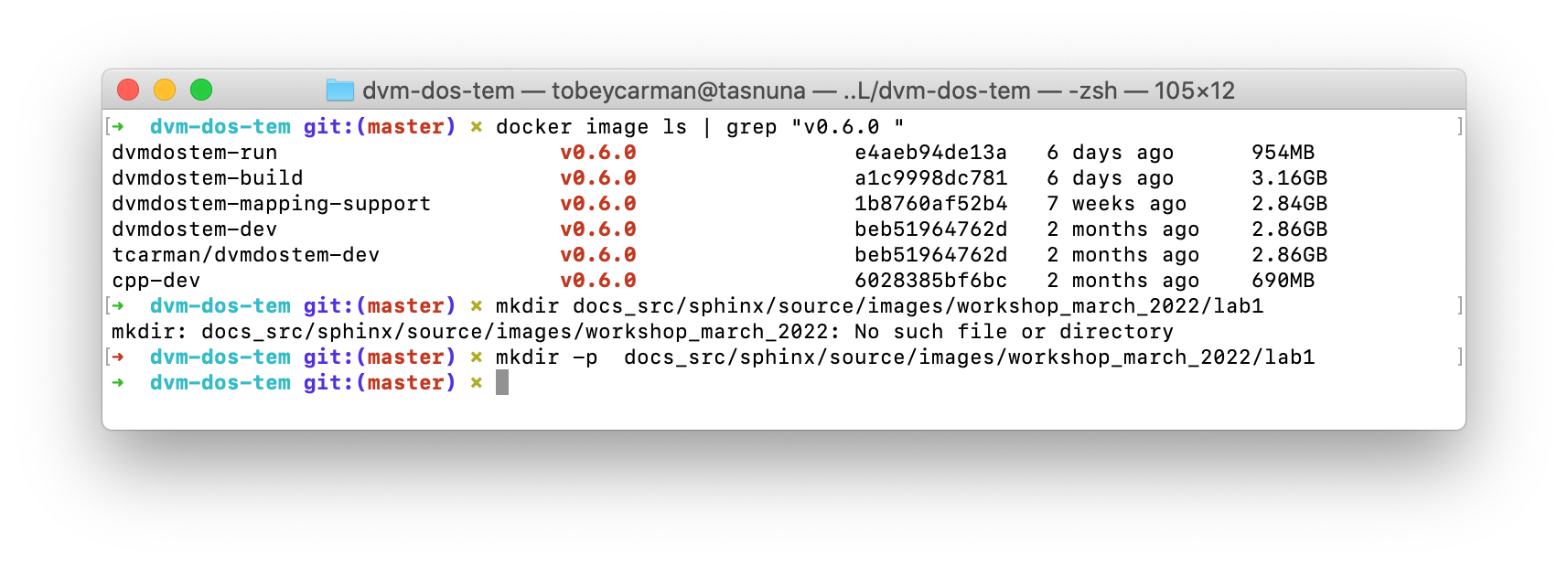
Building the base image, especially cpp-dev, requires quite a bit of downloading and can take 15 minutes or more depending on your internet connection.
When you have built all the images, you should be able to see them in Docker Desktop or with the command line as shown in the screenshot.
2.4. Start and run Docker containers
There are several ways to run a Docker container. The most basic is to use the
docker run command. There are lots of options to this command and it becomes
tedious to provide the options every time you launch the containers. Also some
of the options are the same between different containers. To address this
problem we are using a tool called docker-compose which is bundled with Docker
in recent versions. From the Docker website:
Compose is a tool for defining and running multi-container Docker applications. With Compose, you use a YAML file to configure your application’s services. Then, with a single command, you create and start all the services from your configuration. To learn more about all the features of Compose, see the list of features.
In particular the problem that docker-compose will help us with is mounting
volumes. Volumes provide a way to share data between the host machine (your
computer) and the running containers. Volumes also allow data to persist outside
a container when a container is stopped or shutdown.
Note that in addition to mapping the source code into the containers, we have
also mapped in volumes for the input data and the model output. This means that
on your host machine you need to choose a location for the input catalog and a
location where you would like to store the model output. Once you have chosen
these locations, go ahead and set the environment variables
DDT_INPUT_CATALOG and DDT_WORKFLOWS in a special file named .env
which you need to create in the root of the dvm-dos-tem folder. The
directions for this are at the top of the docker-compose.yml file. Using
this file allows each user to have their own custom locations on their machines
for inputs, outputs, and source code, but inside the containers, the paths are
standardized.
Note
Note that in the design of the Docker images for this project, the
dvm-dos-tem source code is not actually provided inside the image (or
container). The image (and resulting container) only contains the dependencies
and tools for running the code. Thus the source code must somehow be made
available inside the container for any work to be done. This is accomplished
by mounting a volume into the containers when they start. The mounted volume
gives us the ability to share the source code located on your host computer -
the directory that you cloned from Github and have been working with so far in
this tutorial - with the internal run-time environment of the container. We
will also use this tactic to share inputs with the running containers and to
save outputs from the model so that they are available once the container
shuts down.
If you inspect the docker-compose.yml file you will see that there is a
section for each of our containers (called a “service” in docker-compose), and
a section that specifies the volumes. A volume may be mounted in more than one
container or service. For example the volume named “sourcecode” is
specified to use the current working directory on your host with this line:
device: '${PWD}'. Then if you look at one of the containers,
dvmdostem-build for example, you will see that the volume named
“sourcecode” is mapped to the path /work inside the container. This
means that the files on your host computer will be seamlessly linked to the
files inside the container. So for example if you were log in to the container
and create a file named “/work/junk.txt” with some text in it, you should
see that file appear on your host computer at
/path/to/wherever/you/cloned/dvm-dos-tem/junk.txt. This is very powerful
because it allows you to use some of the tools on your host machine to modify
code within the container. For example you can use your text editor of choice
(Sublime, or VSCode, or Notepad++) on your host machine without needing to
install it inside the container!
Note
Currently the semi-automated scripts to generate dvmdostem inputs are very
platform specific and not easy to run. So we have created inputs for about 180
sites across Alaska and can provide them for running dvmdostem. For this
tutorial, it is assumed that you have at least one of these input datasets in
a location on your computer and have set the DDT_INPUT_CATALOG environment
variable to this location.
To launch the containers, use the following command:
$ docker compose up -d
You should get something like the following, and then running $ docker ps
you should see that some of the containers are running. For our use case, we do
not need the cpp-dev or the dvmdostem-build containers to keep running.
They exit immediately, and that is OK.
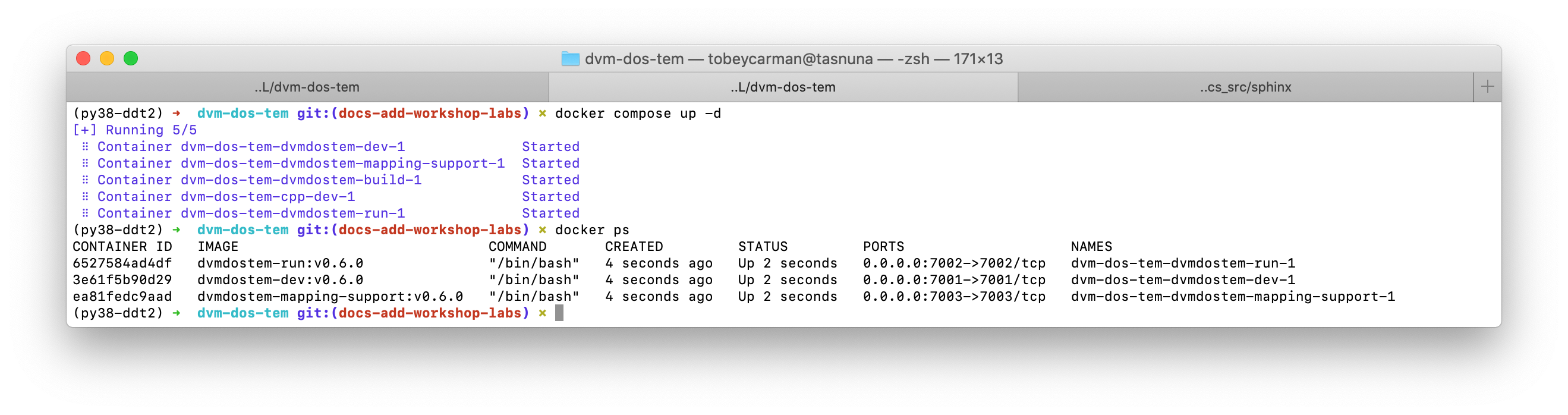
Note that docker compse up with no additional arugments will start all the
containers specified in the compose file. If you wish you can bring up specific
services by naming them on the command line, i.e. docker compose up -d
dvmdostem-autocal, which will start only the auto-calibration service.
With a running container, the most basic thing you can do is log in and poke around. Try this now by running:
$ docker compose exec dvmdostem-dev bash
Which will give you a bash shell inside your container, looking something like this:
develop@ef7aad33441c:/work$
Take some time to poke around. Change directories. List the files. Notice that
you are in the /work directory which is mapped to be your repository folder
on your host machine. Make a new file and see that you can find it on your host.
Take a look at the /data directory and notice that the input catalog and
workflow directory are mapped (linked) to the appropriate directories on your
host machine.
The last step before we can start setting up our model run is to compile the dvmdostem source code. To do this, run the following command:
$ docker compose exec dvmdostem-dev make
... lots and lots of output ...
g++ -o dvmdostem -I/usr/include/jsoncpp obj/ArgHandler.o obj/TEMLogger.o
obj/CalController.o obj/TEMUtilityFunctions.o obj/Climate.o
obj/OutputEstimate.o obj/Runner.o obj/BgcData.o obj/CohortData.o obj/EnvData.o
obj/EnvDataDly.o obj/FireData.o obj/RestartData.o obj/WildFire.o
obj/DoubleLinkedList.o obj/Ground.o obj/MineralInfo.o obj/Moss.o obj/Organic.o
obj/Snow.o obj/SoilParent.o obj/Vegetation.o obj/CohortLookup.o obj/Cohort.o
obj/Integrator.o obj/ModelData.o obj/Richards.o obj/Snow_Env.o obj/Soil_Bgc.o
obj/Soil_Env.o obj/SoilParent_Env.o obj/Stefan.o obj/CrankNicholson.o
obj/tbc-debug-util.o obj/Vegetation_Bgc.o obj/Vegetation_Env.o obj/Layer.o
obj/MineralLayer.o obj/MossLayer.o obj/OrganicLayer.o obj/ParentLayer.o
obj/SnowLayer.o obj/SoilLayer.o obj/TemperatureUpdator.o obj/TEM.o -I/usr/lib
-lnetcdf -lboost_system -lboost_filesystem -lboost_program_options
-lboost_thread -lboost_log -ljsoncpp -lpthread -lreadline -llapacke
which will use the environment and tools inside the container to compile the C++
source code (which is linked into the container via the mounted volume) into the
dvmdostem binary. This can take several minutes. Once it is done you should
have a new file in your repository folder named dvmdostem. You should not
need to run this again unless you modify the C++ source files.
Finally with all this setup in place we can start working on setting up a model run.
2.5. Setting up a dvmdostem run
In general the steps to making a dvmdostem run are as follows:
Decide where on your computer you want to store your model run(s).
Decide what spatial (geographic) area you want to run.
Decide what variables you want to have output.
Decide on all other run settings/parameters:
Which stages to run and for how many years.
Is the community type (CMT) fixed or driven by input vegetation.nc map?
For which stages should the output files be generated and saved?
Calibration settings if necessary (
--cal-mode).Any other command line options or environment settings.
Launch the run.
Verify run completed.
Make plots or other analysis.
The rest of this tutorial will walk through the above steps, doing a very basic
dvmdostem run using the Docker stack.
Note
There are two distinct ways to run commands in the Docker containers:
Interactive - With an interactive command you start by running a one-off command into the Docker container, but the command you run is a shell (Read Eval Print Loop; REPL). With this shell running inside the container you can execute any sort of program that is installed in the container; when the program exits, you are returned to your shell prompt inside the container.
$ docker compose exec dvmdostem-dev bash develop@ef7aad33441c:/work$ ls /data input-catalog workflows develop@ef7aad33441c:/work$ exit exit $
One-off commands - With a one-off command, you execute the command inside an already running docker container (using
docker execordocker compose exec) and when the command is finished, you are returned to the shell on your host computer.$ docker compose exec dvmdostem-dev pwd /work $
Both methods will be used in this tutorial. The different methods can be used to leverage the shell’s tab-complete functionality in different circumstances.
2.5.1. Setting up the working directory
First we are going to set up a working directory where we will conduct our model
run and save the outputs. We will keep this directory inside the workflows
folder (which you linked from your host to the container during the setup
above). There is a helper script for setting up a working directory. This script
will copy over the required parameter and settings files, set up an output
folder and make some adjustments to the configuration file for the run.
Using the one-off command style, run the script. For this case we just arbitrarily select a dataset from your input catalog. Don’t worry if you have a different dataset from the example shown here. If you don’t have any input datasets in your input-catalog, then use the demo-data that is included with the repository.
$ docker compose exec dvmdostem-dev scripts/setup_working_directory.py \
/data/workflows/basic_model_run \
--input-data-path /data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10
Namespace(copy_inputs=False,
input_data_path='/data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10',
new_directory='/data/workflows/basic_model_run', no_cal_targets=False)
which will create a new folder (named basic_model_run) inside your workflows
directory. This folder will have the dvmdostem default parameters copied in
as well as config settings. The paths in the config.js file should be
correctly set to the input data set you chose with the --input-data-path
command line option.
Note
What is with the nonsense that is printed out to your terminal when running various dvmdostem scripts? All of our scripts are essentially rough-draft, so we just haven’t had time to refine the information that is printed out to the console. So when you see stuff like
...
Namespace(copy_inputs=False, input_data_path='/data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10',
new_directory='/data/workflows/basic_model_run', no_cal_targets=False)
Sometimes it is useful and sometimes it isn’t. In most cases it is simply leftover from whatever was needed when the script was developed.
Note
What happens when you get something like this:
$ docker compose exec dvmdostem-dev scripts/setup_working_directory.py \
/data/workflows/basic_model_run --input-data-path \
/data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10
Namespace(copy_inputs=False, input_data_path='/data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10', new_directory='/data/workflows/basic_model_run', no_cal_targets=False)
Traceback (most recent call last):
File "scripts/setup_working_directory.py", line 82, in <module>
shutil.copytree(os.path.join(ddt_dir, 'config'), os.path.join(args.new_directory, 'config'))
File "/home/develop/.pyenv/versions/3.8.6/lib/python3.8/shutil.py", line 554, in copytree
return _copytree(entries=entries, src=src, dst=dst, symlinks=symlinks,
File "/home/develop/.pyenv/versions/3.8.6/lib/python3.8/shutil.py", line 455, in _copytree
os.makedirs(dst, exist_ok=dirs_exist_ok)
File "/home/develop/.pyenv/versions/3.8.6/lib/python3.8/os.py", line 223, in makedirs
mkdir(name, mode)
FileExistsError: [Errno 17] File exists: '/data/workflows/basic_model_run/config'
In this case the error has to do with the directory you are trying to create already existing. This might happen because you ran the script once, then decided to change a setting and tried running it again. Again, most of our scripts are rough-draft and we have not figured out how to gracefully handle all errors yet. It is your responsibility as a user to make sure that the commands complete correctly and if they don’t, to read the traceback and try to figure out what is going on. If you encounter errors and ask a programmer for help, the first thing they will want to see is the command you ran, the error message(s) and the traceback.
To fix the error in this traceback, you would need to delete the offending
directory and run the setup_working_directory.py script again. Or choose a new
directory name.
Let’s look around in the directory you just created.
docker compose exec dvmdostem-dev bash
develop@ef7aad33441c:/work$ cd /data/workflows/basic_model_run/
# Check the files that should have been created with the setup script
develop@ef7aad33441c:/data/workflows/basic_model_run$ ls
calibration config output parameters run-mask.nc
The idea is that each run will exist in its own self-contained directory with
all the config files necessary to execute the run. The output data will also be
stored here. This ensures that the run can be easily adjusted, re-run, and
archived for later use without losing any provenance data. By default the
driving input data is not copied to the experiment folder (to save space). If
you need to copy the driving input data into your experiment directory, try the
--copy-inputs flag.
If you inspect the config/config.js file, you will see that the paths to the
input data are absolute (starting with / and pointing toward the input
dataset that you specified) and the paths to the parameters, run-mask.nc,
calibration folder, outspec.csv and output folder are relative (no leading
/).
2.5.2. Adjusting the config file
Note
Notice that for this run, we only care to run a single pixel (a “site run”) so
why have we chosen a 10x10 pixel dataset (as evidenced by the input dataset
name: ...CALM_Chevak_10x10)? Well our input preparation scripts use GDAL
to extract data from georeferenced .tifs that were created by
https://uaf-snap.org. GDAL’s warping tool can’t create super small grids that
are appropriately geo-referenced. So we have made all our input datasets 10
pixels by 10 pixels (or larger) and then the end user can disable any pixels
they wish by using the run-mask.nc file.
For this totally arbitrary run, let’s turn on outputs for all run-stages (except
pre-run). For more information on what “run stages” are, see here. So open the config/config.js file and
make sure that the following are all set to 1. You can do this with an editor on
your host machine or using vim from inside the container:
"IO": {
...
"output_nc_eq": 1,
"output_nc_sp": 1,
"output_nc_tr": 1,
"output_nc_sc": 1
...
}
2.5.3. Adjusting the run mask
Now let’s adjust the run-mask so that we only run 1 or 2 pixels. Note that you
can use the --show option to see what the mask looks like before and after
adjusting it. We’ll turn on 2 pixels here, just for fun:
# First make sure all pixels are OFF (set to 0)
$ docker compose exec dvmdostem-dev runmask.py --reset /data/workflows/basic_model_run/run-mask.nc
Setting all pixels in runmask to '0' (OFF).
# Then turn one pixel.
$ docker compose exec dvmdostem-dev runmask.py --yx 0 0 /data/workflows/basic_model_run/run-mask.nc
Turning pixel(y,x) (0,0) to '1', (ON).
# And another pixel
$ docker compose exec dvmdostem-dev runmask.py --yx 1 1 /data/workflows/basic_model_run/run-mask.nc
Turning pixel(y,x) (1,1) to '1', (ON).
Note that you don’t want to pass --reset to the second call, or it will
disable the first pixel you enabled!
2.5.4. Choosing the outputs
Next we need to enable some output variables. The control for which outputs
dvmdostem will generate and at what resolution happens using a special
.csv file. The file has one row for every available variable and columns for
the different resolutions. The file can be edited by hand, but we have also
written a utility script for working with the file. We’ll use the utility script
here. For this example we will do our command using the interactive form instead
of the one-off form. Also notice that this script outputs a summary of the
variables enabled in a tabular format. This means that it is hard to read on a
narrow screen because the lines wrap, which is why the following looks so bad.
On your computer you can make the font smaller or your terminal wider to have
the output be readable.
# Get a shell on the container
$ docker compose exec dvmdostem-dev bash
# Change into our working directory for this experiment
develop@ef7aad33441c:/work$ cd /data/workflows/basic_model_run/
# Turn on RH
develop@ef7aad33441c:/data/workflows/basic_model_run$ outspec.py config/output_spec.csv --on RH y layer
Name Units Yearly Monthly Daily PFT Compartments Layers Data Type Description
RH g/m2/time y invalid invalid invalid l double Heterotrophic respiration
# Turn on VEGC
develop@ef7aad33441c:/data/workflows/basic_model_run$ outspec.py config/output_spec.csv --on VEGC m pft
Name Units Yearly Monthly Daily PFT Compartments Layers Data Type Description
VEGC g/m2 y m invalid p invalid double Total veg. biomass C
Warning
The order of arguments to util/outspec.py is very counterintuitive! The
file you want to modify needs to be the first argument so that it doesn’t get
confused with the resolution specification.
Note
Try the --help flag for more options, inparticular, the -s flag for
summarizing the current file.
Every output variable can be produced for a set of dimensions. These dimensions are a reflection of the structure of the model and vary between output variables. The more dimensions are selected, the more information you will get, and the larger the output files will be. For regional runs, a trade-off between the granularity of the outputs needed and the size of the output files needs to be considered.
Three time dimensions are available: yearly, monthly and daily. Daily outputs are only available for a few physical variables, and aren’t generally produced as (1) the model is primarily designed to represent ecological dynamics on a monthly basis, and (2) the amount of data created rapidly becomes unmanageable for multi-pixel runs.
Two dimensions are specific to the vegetation: PFT, i.e. plant functional type, and Compartment. By default, all output variables associated with the vegetation are produced for the entire ecosystem (community type). But every community type is defined by an ensemble of plant function types, which are composed of single or multiple species sharing similar functional traits (e.g. “deciduous shrubs”, “evergreen trees”, “sphagnum moss” …). Finally, every PFT is partitioned into multiple compartments: “leaves”, “wood” (stem and branches), and “roots” (coarse and fine). By selecting PFT and/or compartment, the outputs for a vegetation-related variable will be partitioned by these dimensions.
One dimension is specific to the soil column: layer. The soil column is divided
into multiple layers, that can belong to five types of horizons – brown moss,
fibric organic, humic organic, mineral and rock. By default, soil-related
variables will be summed-up across the entire soil column. But if the layer
dimension is selected in the output_spec.csv file, the selected variable
will produce ouputs by layer.
2.6. Launch the dvmdostem run
Finally we are set to run the model! There are a number of command line options
available for dvmdostem which you can investigate with the --help flag.
The options used here are for setting the length of the run-stages, for controlling the log level output, and for
forcing the model to run as a particular community type.
In a real run --eq-yrs might be something like 1500 and --sp-yrs
something like 250. But for testing we might be too impatient to wait for that.
Plus for this toy example, we enabled fairly hi-resolution outputs so running
the model for long time spans could result in huge volumes of output. The
dvmdostem model is fairly flexible with respect to outputs and output
resolution so the user must put some thought into choosing configurations that
make sense and are reasonable for the available computing power.
In this case we are forcing the community type to be CMT 4. In a “normal”
dvmdostem run, the community type is controlled by the input
vegetation.nc file. This file has a CMT code for each pixel, which
corresponds to the CMT numbers in the parameter files. Frequently for single
pixel runs the user wants to ignore the vegetation.nc map and force the
pixel to run as a particular CMT. In this case we want to force our pixel to run
as CMT 4 simply because we know that the parameters for CMT 4 have been
calibrated. For more discussion about community types in dvmdostem see the
CMT section.
The --log-level command line option controls the amount of information that is
printed to the console during the model run. There are 5 levels to choose from:
debug, info, note, warn, error, fatal. With debug level, all print statements in
the model code are enabled and the output is extremely volumnious, but useful
for tracking down issues with the code. With the fatal level, only a small
handful of messages will be printed out. This is useful for production runs, but
if a run fails, it can be difficult to know why.
Note
Despite the fact that there is a command line option for pointing to an
arbitrary control file (config.js), this option doesn’t work when used with
relative paths in the control file as we have for this lab. For this reason we
provide the --workdir /data/workflows/basic_model_run/ command line option
when launching the model. Notice that this command line option is associated
with docker compose exec, not dvmdostem.
Note
The organization of the log messages is not complete and is actually quite
messy. So for example you will find that when running with --log-level err you
will get lots of mundane messages noting the year or other non-error things,
e.g.:
...
[fatal] [EQ] Equilibrium Initial Year Count: 5
[fatal] [EQ] Running Equilibrium, 5 years.
[err] [EQ->Y] y: 0 x: 0 Year: 0
[err] [EQ->Y] y: 0 x: 0 Year: 1
[err] [EQ->Y] y: 0 x: 0 Year: 2
[err] [EQ->Y] y: 0 x: 0 Year: 3
...
This is simply because we have not gone through the code base and re-categorized all the messages. Until this is fixed, you simply have to experiment with the different levels until you are seeing the output that is appropriate for your particular use-case.
Launch the model with the following command, note that the majority of the console output has been omitted for clarity:
$ docker compose exec --workdir /data/workflows/basic_model_run/ dvmdostem-dev dvmdostem -l err -f /data/workflows/basic_model_run/config/config.js -p 50 -e 100 -s 25 -t 115 -n 85
Setting up logging...
[err] [] Looks like CMTNUM output is NOT enabled. Strongly recommended to enable this output! Use outspec.py to turn on the CMTNUM output!
[err] [PRE-RUN->Y] y: 0 x: 0 Year: 0
[err] [PRE-RUN->Y] y: 0 x: 0 Year: 1
[err] [PRE-RUN->Y] y: 0 x: 0 Year: 2
...
[err] [PRE-RUN->Y] y: 0 x: 0 Year: 49
[fatal] [EQ] Equilibrium Initial Year Count: 100
[fatal] [EQ] Running Equilibrium, 100 years.
[err] [EQ->Y] y: 0 x: 0 Year: 0
[err] [EQ->Y] y: 0 x: 0 Year: 1
...
[err] [SC->Y] y: 1 x: 1 Year: 82
[err] [SC->Y] y: 1 x: 1 Year: 83
[err] [SC->Y] y: 1 x: 1 Year: 84
cell 1, 1 complete.35
[fatal] [] Skipping cell (1, 2)
[fatal] [] Skipping cell (1, 3)
...
[fatal] [] Skipping cell (9, 7)
[fatal] [] Skipping cell (9, 8)
[fatal] [] Skipping cell (9, 9)
Total Seconds: 70
With a quick glance at the console output, it looks like the run completed without problems. We can further verify this by looking at the run-status.nc file in the output folder, for example:
$ docker compose exec dvmdostem-dev ncdump /data/workflows/basic_model_run/output/run_status.nc
netcdf run_status {
dimensions:
Y = 10 ;
X = 10 ;
variables:
int run_status(Y, X) ;
data:
run_status =
100, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 100, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0 ;
}
A positive status code means the pixel completed, while a negative status code
indicates that the pixel failed for some reason. In the event of a pixel
failing, there will be some kind of error messages in a fail_log.txt file in
the output folder. In this case it appears that both our pixels completed
successfully. The next step is digging into the output data to see what it looks
like.
Note
To see what would happen if we did not provide the --force-cmt command
line option, we need to investigate the vegetation.nc input file, and
specifically for the two pixels we have enabled (0,0) and (1,1). The path to
the file is in the config/config.js file. We can use grep to find this
line:
$ docker compose exec dvmdostem-dev grep vegetation.nc /data/workflows/basic_model_run/config/config.js
"veg_class_file": "/data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10/vegetation.nc",
From here there are many ways we could go, but for this example we will use the command line ncks (netcdf kitchen sink) tool to print out the variable from the file:
$ docker compose exec dvmdostem-dev ncks -v veg_class \
/data/input-catalog/cru-ts40_ar5_rcp85_ncar-ccsm4_CALM_Chevak_10x10/vegetation.nc
netcdf vegetation {
dimensions:
X = 10 ;
Y = 10 ;
variables:
char albers_conical_equal_area ;
albers_conical_equal_area:grid_mapping_name = "albers_conical_equal_area" ;
albers_conical_equal_area:false_easting = 0. ;
albers_conical_equal_area:false_northing = 0. ;
albers_conical_equal_area:latitude_of_projection_origin = 50. ;
albers_conical_equal_area:longitude_of_central_meridian = -154. ;
albers_conical_equal_area:standard_parallel = 55., 65. ;
albers_conical_equal_area:longitude_of_prime_meridian = 0. ;
albers_conical_equal_area:semi_major_axis = 6378137. ;
albers_conical_equal_area:inverse_flattening = 298.257222101 ;
albers_conical_equal_area:spatial_ref = "PROJCS[\"NAD83 / Alaska Albers\",GEOGCS[\"NAD83\",DATUM[\"North_American_Datum_1983\",SPHEROID[\"GRS 1980\",6378137,298.2572221010002,AUTHORITY[\"EPSG\",\"7019\"]],AUTHORITY[\"EPSG\",\"6269\"]],PRIMEM[\"Greenwich\",0],UNIT[\"degree\",0.0174532925199433],AUTHORITY[\"EPSG\",\"4269\"]],PROJECTION[\"Albers_Conic_Equal_Area\"],PARAMETER[\"standard_parallel_1\",55],PARAMETER[\"standard_parallel_2\",65],PARAMETER[\"latitude_of_center\",50],PARAMETER[\"longitude_of_center\",-154],PARAMETER[\"false_easting\",0],PARAMETER[\"false_northing\",0],UNIT[\"metre\",1,AUTHORITY[\"EPSG\",\"9001\"]],AUTHORITY[\"EPSG\",\"3338\"]]" ;
albers_conical_equal_area:GeoTransform = "-613280.1453076596 1000.050170338725 0 1346667.58012239 0 -999.7969847651308 " ;
int veg_class(Y,X) ;
veg_class:grid_mapping = "albers_conical_equal_area" ;
data:
albers_conical_equal_area = "" ;
veg_class =
4, 4, 4, 4, 4, 4, 0, 4, 4, 4,
4, 4, 4, 6, 6, 4, 4, 4, 4, 4,
4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
4, 4, 4, 4, 4, 4, 4, 4, 4, 4,
4, 4, 4, 4, 4, 4, 4, 4, 4, 6,
4, 4, 4, 4, 4, 4, 4, 4, 6, 4,
4, 4, 4, 4, 4, 4, 4, 6, 0, 4,
6, 4, 6, 4, 4, 4, 4, 4, 4, 4,
6, 6, 6, 4, 4, 4, 4, 4, 4, 4,
6, 6, 6, 4, 4, 4, 4, 4, 4, 4 ;
} // group /
This output is a little bit ugly to read, mostly due to the metadata that
addressed the geo-referencing. But if we skim past all the metadata, we find
the actual data array for the veg_class variable. Because the array is
only 10x10, the printed output is readable and we can see that incidentally,
pixels (0,0) and (1,1) are set to CMT 4. So it turns out that the
--force-cmt isn’t doing anything in this case. Oh well!